Author: Dr Mac-Edwin Ifeanyi Obi (mobi184@aiu.edu.gm, +220 786 1972)
COVID-19, otherwise known as coronavirus disease was first reported on December 27, 2019 in Wuhan, Province of Hubei, Republic of China. At the time of writing this paper, May 15, 2020, over three million persons have tested positive for SARS-CoV-2, the virus that causes COVID19. About 10% of the infected has died. The World Health Organisation (WHO) rightly declared the outbreak a pandemic on March 11, 2020. As a result, most people in the world are living under some form of restriction.
Little research work has been done on SARS-CoV-2 as it is a new virus. This has made controlling the COVID-19 pandemic challenging. A better understanding of the unique features of the virus and clinical presentation of COVID-19 patients is necessary to develop accurate and fast diagnostic tools for COVID-19. Hence, a need for this research work.This research involves searching medical journal for works on diagnostic tools for COVID-19 and critically appraising them for evidence. This must be done before they can be applied clinically.
21,269 titles were scanned through and 20 selected papers were reviewed. The first paper is prediction models drawn up with clinical features of fever, (dry) cough and shortness breath alongside chest CT Scans. The researchers posted high C indices of 0.78-0.81 which indicate high statistical significance. Two other studies gave RT-PCR along with chest CT Scans as the gold standard for diagnosing COVID-19. A study put the sensitivity of chest CT Scans at 97% (CI 95-98%; p-value < 0.001). Most studies at this stage on COVID-19 are at high risk of bias as study participants are severely sick and sample sizes are small.
Keywords: COVID-19, SARS-CoV-2, RT-PCR, viral genome
As at June 15, 2020 over 8 million individuals have tested positive for SARS-CoV-2, the viral pathogen of COVID-19. The outbreak has assumed a pandemic trend and nothing has caused more disruption to life as the pandemic has since World War II.
There is need for rapid and accurate diagnosis of the disease. Several researchers have done one study or the other in order to come up with the most sensitive and specific diagnostic tool to identify the virus; and by extension, diagnose the disease. There has been prediction models, laboratory guideline and protocols, molecular tests, serological test and radiological (imaging) scans. Even deep machine learning (artificial intelligence) has been applied to help successfully put the pandemic in check (Vaishya et al., 2020).
COVID-19 is a highly infectious disease caused by SARS-CoV-2. SARS-CoV-2 is a member of the coronaviridae family. Other members of the family include, inter alia, MERS virus, SARSCoV, the pathogen of Severe Acute Respiratory Syndrome which well-known epidemic rocked mainly mainland China and Hong Kong in 2003.

Figure A1- Image of SARS-CoV-2
Adapted from CDC Image Library [Available at www.cdc.gov/imagelibrary ; accessed at May 15, 2020 1815hrs GMT]
Signs and Symptoms of COVID-19
COVID-19 first manifests clinically with the presence of fever, generally 38.5 degree Celsius. Thereafter, dry cough that continues for up to three straight days is observed. Some patients, particularly those with impaired immunity and respiratory ailments usually progress to shortness of breath. This state of inability to breathe normally causes most of the COVID-19 deaths.
Some clinicians have also noted the presence of chest pain, chills, and impaired sense of smell and taste in some COVID-19 patients.
COVID-19 Diagnostics
A good number of these testing procedures has never been empirically proven by clinical research but because of the huge pressure that been brought to bear on the scientific community to control the monster, everything seems to have been thrown into the mix!
The essence of this systematic review and critical appraisal is to go through published (and even preprints) and identify diagnostic procedures that have been proven objectively to deliver quick and effective identification of the causative virus. These evidence-based procedures can then be applied clinically to diagnose COVID-19.
Going further, suggestions will be made on how the other diagnostic tests that have not been independently proven can be enhanced in quality so they can be used clinically. As things stand, what is known and used are clinical symptoms –fever, headache, continuous (dry) cough, myalgia and when severe, shortness of breath. These are basically used as initial screening tests to identify suspected cases. The second level of diagnostic process involves the use of molecular tests, particularly RT-PCR to identify active viral shedding.
The RT-PCR is the main testing modality available at most COVID-19 treatment centres. It involves the amplification of particular DNA sequences of interest so it can be studied.Since the entire genome of the SARS-CoV-2 was made available, molecular processes like RT-PCR can now be done.
1 ctttcgatct cttgtagatc tgttctctaa acgaacttta aaatctgtgt ggctgtcact
61 cggctgcatg cttagtgcac tcacgcagta taattaataa ctaattactg tcgttgacag
121 gacacgagta actcgtctat cttctgcagg ctgcttacgg tttcgtccgt gttgcagccg 181 atcatcagca catctaggtt tcgtccgggt gtgaccgaaa ggtaagatgg agagccttgt
241 ccctggtttc aacgagaaaa cacacgtcca actcagtttg cctgttttac aggttcgcga ……..
Figure A2-Part of the Genome of SARS-CoV-2; the full genome is published on genbank. Adapted from http://www.snapgene.com; accessed May 12, 2020 1345 hrs GMT
Figure A3-Diagram Depicting the RT-PCR Process
Adapted from Google images [Available at http://www.google.com ; accessed May 12, 2020 1304 hrs GMT]
SARS-CoV-2 is an RNA virus and has to be first converted to complementary DNA before it can be replicated. This is done by the enzyme reverse transcriptase. The process involves heating and
cooling specific gene segment until the sequence of interest is isolated and replicated. It can then be studied. RT-PCR takes about 24 hours to be completed.
Radiological tools like chest CT Scans (or X-ray in our under-resourced country) are known to complement the molecular tests result as they show a classic bilateral `ground glass opacity`.
Serological (antibody) tests could provide not only rapid diagnostic tools but can also confer immunity on recovered patient. Research in the future will surely unravel this. This phase of diagnosis is more rapid that RT-PCR which is longer. However, it is challenging as it takes long for antibodies to be developed. Again, recovered patients have to be studied fro a long time before immunity from infection can established. This is the reason why developing a rapid diagnostic test for the Ebola virus has been challenging.
COVID-19 Prophylaxis-The following are preventive measure for COVID-19:
Clean your hand often with soap and water and/or alcohol/based hand sanitizer. 2.Manitain a safe distance of between one to two metres from the person next to you.
Do not touch your eyes, nose or mouth.
Cover your mouth and nose with your bent elbow whenever you cough or sneeze.
Stay home if you feel unwell. If you have fever, cough and/or difficulty breathing call your local health authority. Follow all the directives of your local health authority. 1.4.2 Prospects of Vaccine Development-About ninety vaccines are at different stages of development and some have got to clinical trial stage. Preliminary studies from patients who recovered from COVID-19 have shown antibodies to SARS-CoV-2. The other coronavirus outbreaks of Middle East Respiratory Syndrome (MERS) and Severe Acute Respiratory Syndrome (SARS) were quickly quashed or did not cause much global distress and as such no vaccines were developed for them. Antibodies were detected in recovered COVID-19 patients and this is the basis of vaccine development and further research in future will make things clearer.
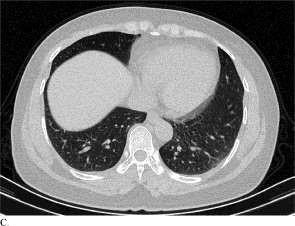
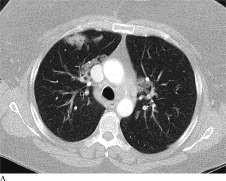
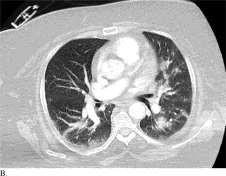
Figure C is from a patient with COVID-19; Figures A and B are from patients with non- COVID19 viral pneumonia
Adapted from Bai et al., [Available at .
PubMed Central, bioRxiv and Google Scholar were searched on April 18, 2020. While PubMed gave 1,248 titles; bioRxiv and Google Scholar gave ten more titles. A good number of the titles from bioRxiv and Google Scholar were also seen on PubMed Central. Over the following week, PubMed Central was searched further and this time, the search was modified with `COVID-19` or `Coronavirus Diagnostics`. This yielded over 21, 269 titles. Most of the 1,248 titles I got in my earlier search were in the later one. I realized that the 1,248 titles were the most specific for diagnosis of COVID-19. The larger (21,269) titles contains origin, epidemiology, pathogenesis, treatment and prognosis in addition to diagnosis. I then settled for the earlier search (1,248 titles).
The captions and abstracts of the 1,248 titles were scanned through to arrive at a final 20 most relevant papers. All 20 papers were published online in English (or had English translation). The studies were conducted in diverse COVID-19 hotspots- China, the US, Germany, Italy, Japan, Denmark and Canada. One paper published in The Gambia traced the viral strains isolated from the cases in The Gambia to Japan, and England and Spain (Europe) (Kanteh et al., 2020).The paper was not reviewed as it was published after I had concluded my review.
3.2 Methods (including Study Design)
3.2.1 Study Design-Of the selected 20 papers, two were `rapid` systematic reviews and critical appraisals. Two randomised clinical trials and sixteen nonrandomised clinical trials. Data was
extracted after thoroughly reading the papers and assessing for risk of bias (ROB), effect of confounding factors, clinical and statistical significance, error and generalisability of the research findings.
To test for the ROB, the population studied was evaluated for evidence of selection or observer bias. Was it made up of individuals of a particular demographic class? Where they only elderly or just the young? The patients` health status at the time they were recruited for the study was evaluated as well. An ideal study ought to have the right mix of patients of varying states of health.
3.2.2 Data Analysis-The time-tested method of controlling confounding factors-age-matching, blinded, randomised clinical process with stratification and multiple variable analyses were looked for in the papers. 95% Confidence Interval (CI) and C-Statistical Index (C Index) were my measures of statistical significance. One of the reviewed papers looked at calibration of prediction model as a measure of accuracy of findings of the study (Gong et al., 2020). Error was measured by the p-value (see list of abbreviations/glossary). Very few of the reviewed papers gave the p-values of their results and they were used to assess for error due to chance.
The papers were critically appraised by looking at the clarity of the study question, internal validity, statistical significance and external validity of the results. For internal validity, the risk of bias and efforts by the researchers to control confounding factors were assessed. The p-value and confidence intervals were studied to determine the statistical significance of the values reported at the end of the studies. The generalisability of the results of the studies was used as a measure of the external validity of the studies` results.
21,269 titles were retrieved from searches conducted on April 18, 2020, April 26, 2020 and May 2, 2020. Of the 21,269 papers, 30 papers were analysed and utilised for the abstract of this paper. Subsequently, 20 studies met the inclusion criteria for systemic review and critical appraisal:
(Wynants et al., 2020), (Vaishya et al., 2020), (Chan et al., 2020), (Bai et al., 2020), (Ai et al., 2020), (Kooraki et al., 2000), (Xia et al., 2020), (Qin et al., 2020), (Kang et al., 2020), (Sigrist et al., 2020), (Konrad et al., 2020), (Shang et al., 2020), (Kai, H, Kai M, 2020), (Stefanelli et al.,
2020), (Holland et al., 2020), (Rothe et al., 2020), (Chen et al., 2020), (Xu et al., 2020), (Metsy
et al., 2020), and (Reed SE, 2020). 4.1 Primary Data
Two rapid reviews, two randomised clinical trials, sixteen nonrandomised clinical were critically appraised. The grouping of the studies clinical trials into randomised and non-randomised was my idea. The researchers did not specifically mention randomization. I came to the conclusion after looking at the features of the studies.
The two rapid reviews were done in Belgium, The Netherlands and India. Two randomised clinical trials were done in China and the US. The nonrandomised trials were done in China, Japan, Germany, Italy and the US. The rapid reviews and critical appraisal were on prediction models for COVID-19 diagnosis and application of artificial intelligence in diagnosing COVID- 19.
Review 1-Prediction Models for Diagnosis and Prognosis of COVID-19 Infection-Systematic Review and Critical Appraisal
31 models from 27 studies were reviewed by Laure Wynants and colleagues. The data for the studies were collected between December 8, 2019 and March 15, 2020. There is a very high chance of overlap as several researchers got data from a single Chinese source.
The median follow up period of most studies were not reported but one paper gave 8.4 days (Dong et al., 2020) and another, 15 days (Shi et al., 2020).
Availability of Models in Format for Use in Clinical Practice
Twelve studies were in a form in which they can be used a clinical screening tool.
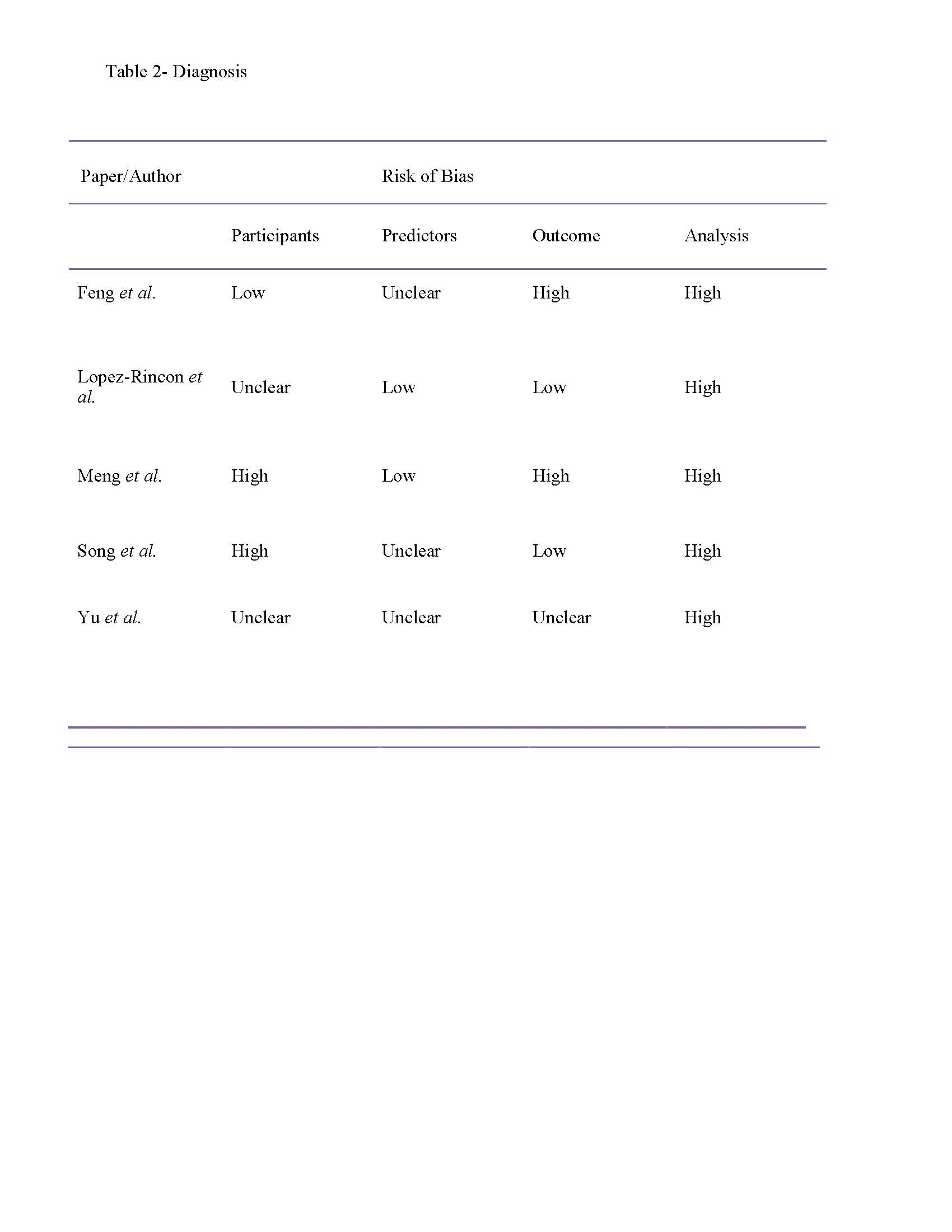
The 20 papers were critically appraised for internal validity, statistical significance, chance (error) and external validity. For internal validity, bias and control of confounding factors were checked. Most of the twelve models are at a very high risk of bias, particularly selection and nay, observer bias. This was picked up by the use of the PROBAST tool, a statistical tool that identifies bias (see table 1). The selection appears to have been done in a haste for researchers to come up with diagnostic tools going by the pressure of the pandemic. Furthermore, there is a risk of confounding as some of the research papers used to draw up the models was developed from
patients with other viral pneumonia, like influenza and cytomegalovirus pneumonia other than COVID-19 pneumonia.
Statistical significance and error again could not be assessed as neither the p-value or confidence intervals of the models were reported.
External validity was not tested as research papers used to develop the models were done in China. Generalisability of the studies can only be assessed when the research is repeated outside where it was originally done under the same conditions.
Models to Predict Risk of Hospital Admission for COVID-19 Pneumonia in General Population Another three models predicted risk of hospital admission for COVID-19 pneumonia.
The predictors of the models are age, sex, previous hospital admission, comorbidities and social determinants of health.
On assessment of internal validity, the studies in these models were at high risk of bias, particularly selection bias. PROBAST was used to assess this. There also was likelihood of confounding as it was not controlled. Data from patients with other viral pneumonia like non-TB pneumonia, influenza, acute bronchitis and upper respiratory tract infections were utilized to develop the model. The use of data from other viral causes surely introduces an error in the analysis of the findings.
Even though the other viral pneumonia have similar clinical presentation as COVID-19, there are subtle differences. In the case of COVID-19, there is the severe shortness of breath that
is not seen in many other viral pneumonia (here, accessed May 13, 2020; 0107 hrs GMT).
Even though the clinical significance of models is in doubt their statistical significance is high (C Indices 073, 0.81 and 0.81).
External validity cannot be assessed as the study has not been repeated outside where it was originally done.

Adapted from Wynants et al., [Available at http://www.bmj.com; accessed May 11, 2020 1800 hrs GMT]
Diagnostic Models to Detect COVID-19 Infections in Patients with Symptoms
One model was able to detect COVID-19 pneumonia in fever clinic patients (estimated C Index 0.94) (Feng, Huang, et al., 2020). Three other models reported: C index of 0.97 suspected COVID-19 patients (Song et al., 2020), C index of 0.87 in asymptomatic patients (Meng et al., 2020) ;and an F1 score of 1.00 which points to 100% sensitivity and specificity in children using symptoms and based on bilirubin and alanine transferase levels (Yu et al., 2020). The predictors
used samples sizes of 2 (patients with fever) to develop the model. These are rather too small to make for statistical significance.
Diagnostic Models Based on Computed Tomography (CT) Imaging
From the report of the researchers, three of the seven models based on positive CT Scans, were artificial intelligence-based. The authors of the review went further to report that one of the models was already in use in sixteen hospitals. However, no mention was made of how it was deployed and success of the models in correctly diagnosing COVID-19. Similarly, the details of the other models were not made available by the researchers.
Other Models
The rest of the models in this review were on the prognosis of COVID-19 patients. The researchers used predictors like age, comorbidity, prior hospitalisation and clinical symptoms. The models achieved high C indices (which signify statistical significance) .But like the other models earlier reviewed, sample sizes were small and there was a high risk of bias as picked up by the PROBAST tool. Having noted the above, prognosis of COVID-19 patients is not within the scope of this systematic review. Therefore, detailed appraisal of the prognostic models was not done and is not presented here.
Review 2- Artificial Intelligence (AI) Applications for COVID-19 Pandemic
The authors of the review looked at seven papers on applying artificial intelligence tools in the control of COVID-19 pneumonia. The researchers, however, failed to give a detailed manner through which they came about their findings (Vaishya et al., 2020). The worksheets used in collecting data were not made available. No mention of measures of statistical significance like C Index or Confidence interval of values was provided.
There is no evidence to apply whatever has been posited by the authors as there is no proof they will work. Further research is definitely needed in this field.
Randomised Clinical Trials
Two randomised clinical trials were reviewed. The first of the trials compared the sensitivity of the regular replication machinery of the SARS-CoV-2, the RdRp antigen protein with the
modified RdRp protein, the RdRp/ Hel. The RdRp/ Hel was synthesised by adding the enzyme, helicase to the regular RdRp.
273 samples were collected from 15 patients admitted at a treatment facility. The samples were collected from nasopharyngeal and nasal cavities. The RdRp antigen protein was extracted. A part of the collected RdRp protein was modified by addition of helicase enzyme.
On comparing the two- RdRp versus RdRp/ Hel, the later recorded a sensitivity of 43.6% (119 out of 273 samples) against the regular RdRp`s sensitivity of 28.3% (77 out of 273). Furthermore, RdRp also successfully detected SARS-CoV-2 in 42 samples which the regular RdRp recorded as negatives (false negatives, that is). The results indicated that RdRp/Hel is far more sensitive in detecting SARS-CoV-2, by extension correctly diagnosing COVID-19. The pvalue of the study is < 0.001 meaning the values are statistically significant.
Going further, internal validity- bias and effect of confounding factors were assessed. The study is at a very high risk of bias, particularly selection bias. The patients were all symptomatic when they were recruited for the study. This means the findings of the study may not apply to asymptomatic carriers of the COVID-19 virus. Then, there is nothing done in the design of the study to show that measures were taken to control for confounding. The clinical symptoms (fever, cough, myalgia and shortness of breath) and the ground glass opacity on chest CT Scan used to recruit the participants of the study are not specific for COVID-19. Therefore, it is possible asymptomatic COVID- 19 cases could have been missed out. This could reduce the positive predictive value of the RdRp/Hel and RdRp and by extension their sensitivities.
Assessment was made to find out if the values were due to chance. A p-value of <0.001 indicates the values were not due chance, they have statistical significance. The study was done in China and there is no report of it being repeated elsewhere. Until that is done, it is not generalisable and external validity cannot be guaranteed.
The second reviewed randomised clinical trial is on the capabilities of five Chinese and American radiologists to interpret CT Scans (Bai et al., 2020). The retrospective cohort study was conducted with 424 participants-205 non-COVID-19 patients from the US and 219 COVID19 patients from seven hospitals in Hunan Province, China between January 6 and February 20, 2020. The non-COVID-19 patients had other viral pneumonia like influenza,
cytomegalovirus pneumonia. The average number of days between positive chest CT-Scans and positive RT-PCR result was 4.1+/-4.4 days. The radiology search engine, MONTAGE, was used to get CT Scans for this study from medical records. The software, Picture Archiving and Communications Systems (PACS) was used to download the selected CT Scans. The Chinese radiologists were blinded to CT Scans, the researchers did not say if the American radiologists were blinded as well.
The results of the study showed that the three Chinese radiologists` accuracy in differentiating COVID-19 from non-COVID-19 pneumonia was 83% (95% CI 79-86%), 80% (95% CI 7683%)
and 60% (95% CI 55-65%). Sensitivity ranged from 72 to 94% while specificity widely varied
from 24 to 94%.
For the Americans, in an age-matched cohort of 58 patients, accuracy were 97% (95% CI 88100%), 88% (95% CI 77-95%), 83% (95% CI 71-91%) and 84% (95% CI 73-93%).
Sensitivity ranged from 70 to 93% and specificity from 93 to 100%. The researchers concluded that the seven radiologists had high specificity and moderate sensitivity in identifying COVID-19 by interpreting chest CT Scans.
Internal validity of the study was assessed by looking for bias and effects of confounding factors. The study was at a high risk of bias. All the COVID-19 positive CT Scans were taken from the Chinese hospitals while the non-COVID-19 CT Scans were from the US. This introduces some selection bias and can affect the final accuracy, sensitivities and specificities results. Having all the COVID-19 CT Scans from China could not guarantee accurate results at the end of the study. The researchers could have taken some COVID-19-positive CT Scans from the US and vice versa.
The researchers made efforts to counter the effects of confounding factors. 58 age-matched controls were used in the study. However, the results of the study would have been a lot more reliable if age-matched controls were used for all the participants.
The 95% Confidence Intervals of the study results indicate they are statistically significant and not due to chance. It could also be deduced from the confidence intervals that the results are fairly precise. The generalisability of the study is doubtful as all COVID-19- positive CT Scans were taken from China and all the COVID-19-negative CT Scans from the US. The results of the
study could change if the reverse was done or if the CT Scans were mixed, that is, some COVID19-positive scans from the US and the other from China and vice versa.
Sixteen nonrandomised clinical trials were reviewed. The trials dwelt on chest CT Scans, molecular and serological diagnostic tests of COVID-19. One interesting study looked at the diagnostic value of saliva. There were doubts in some quarters if the COVID-19 pathogen could be detected in saliva, the results of this study says it could.
Clinical Trials on CT Scans
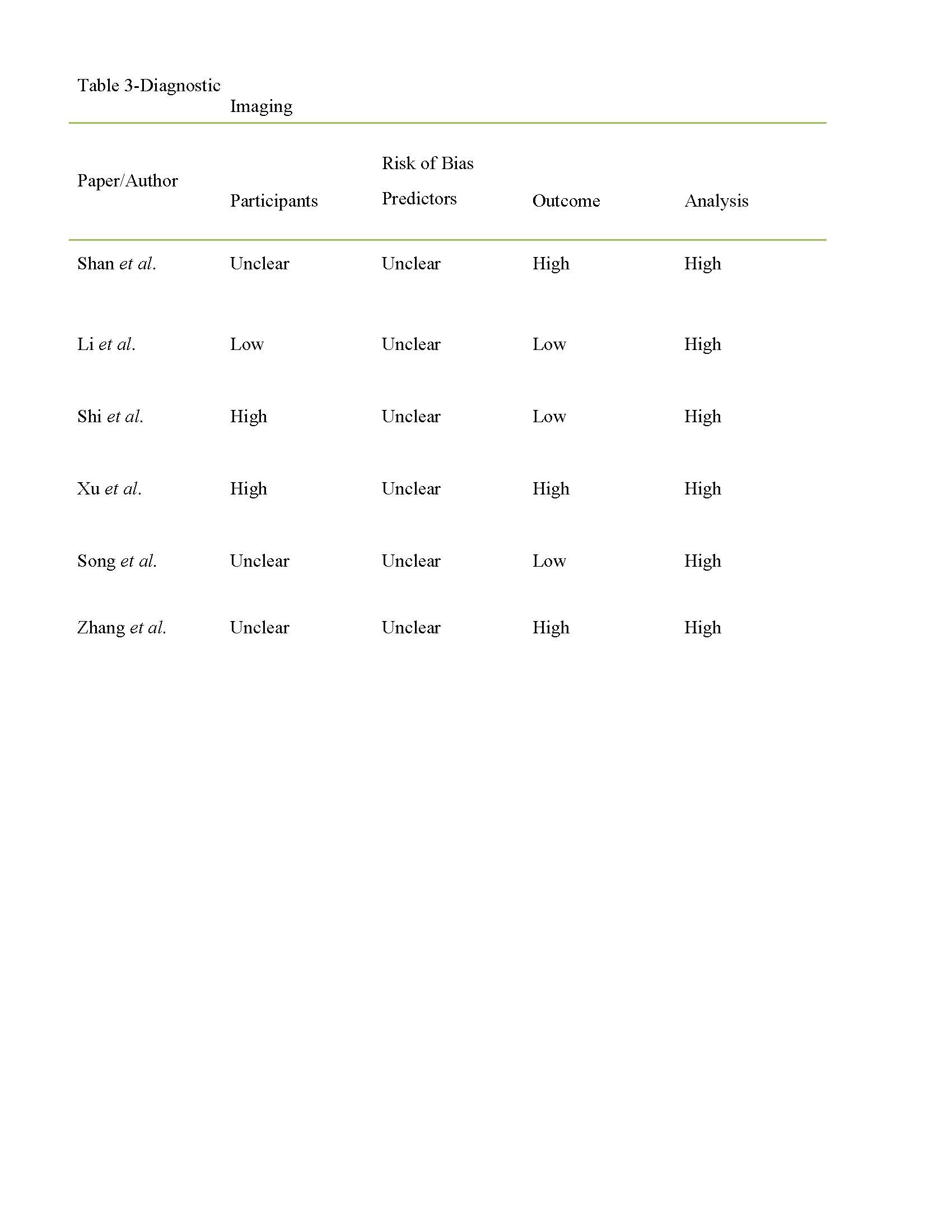
The first nonrandomised clinical trial reviewed was authored by Ai T and his colleagues. The study was done in Wuhan, Hubei Province of China and the study basically compared the sensitivity and specificity of RT-PCR antigen test and radiological investigation (Chest CT Scans) as investigative tests for COVID-19.
Internal validity (bias and confounding risk), error (due to chance) and external validity (generalisability) were evaluated. There is high likelihood of selection bias because the study was done with hospitalized patients and the results will likely be different if asymptomatic patients were included. The values from the study are statistical significance from the 95% confidence interval of the values. Also, the values are very precise. The study is a randomised trial, however it was not mentioned if it was blinded.
However, the p-values were not made available by the researchers. This value, if given could have made the values far more significant and reduce further the probability of the figures being as a result of chance.
The findings cannot be generalised at this stage until repeated in other parts of the world under identical experimental conditions.
Another of the nonrandomised studies on value of CT Scans in diagnosing COVID-19 looked at the chest CT-Scans of patients from Chinese hospitals (Kooraki et al., 2020). The 41 cases had earlier been confirmed with the RT-PCR antigen protein test. The researchers recorded that
radiologists were able to correctly identify COVID-19 in all 41 cases; sensitivity of 100% (Huang C, Wang Y, Li X, 2020).
Internal validity (bias and confounding risks), error (due to chance) and external validity (generalisability) were assessed. The risk of bias is high as only hospitalised were used in the study. The results will likely differ if asymptomatic patients are included. No p-values or confidence intervals were given by the researchers; hence statistical significance cannot be measured. The study was done in Wuhan, China. Unless it is repeated under the same conditions elsewhere, the results of the study cannot be generalised.
Another paper on chest CT Scan features in paediatric patients in Wuhan, Hubei Province, China was reviewed (Xia et al., 2020). The researchers recruited 20 inpatients in a hospital in Wuhan. They were studied for symptoms and CT Scan findings. The researchers reported that 65% of the patients had cough while 60% had fever as presenting clinical features.
Further review for internal validity reveals there is a high risk of bias as all participants were admitted patients. The results would have been a lot more reliable if there were outpatients. There were no controls to nullify effects of confounding factors. Also, the researcher did not give the body temperature that was used as benchmark for fever. It must be noted that different parts of the body gives different temperatures. This is more important in paediatric patients and ought to have been spelt out by the authors.
Confidence intervals and the p-values of the study were not given and as such the statistical significance of the study is not known. The likelihood of the findings being due to chance is high. External validity cannot be assessed as the study has not been repeated outside the hospital where it was originally done in Wuhan, China. The results of the study can therefore not be generalised; thus it cannot be utilised clinically.
The fourth nonrandomised clinical trial on CT Scans was done with four participants who had been confirmed with COVID-19 with `ground glass opacity` on their chest CT-Scans. The trial involved doing a PET scan with the injection of 18-F FDG tracer (Qin et al., 2020). The researchers found out that three of the four patients recorded increased 18-F FDG uptake.
Going further, there is high risk of bias as the modalities of selecting the four participants were not given. Again, they were said to be positive but the test done to confirm their status was not given. A`ground glass opacity on chest CT-Scan is not specific for COVID-19 and could be any other viral pneumonia. There was no comparator so confounding could not be controlled. The pvalue was not given and as such statistical significance cannot be determined. The low sample size also makes statistical significance less likely. The findings of the study cannot be generalised until it is repeated outside the facility where it was done in China.The fifth nonrandomised study about chest CT Scans reviewed is on the use of low-dose chest CT Scans in the detection and management of COVID-19 (Kang et al., 2020). The study population is two COVID-19 patients and they both got 1/9 of the standard chest CT Scan dose (dose length14.5m GY cm, effective dose 0.203mSv) and the standard dose (dose length 129.1m GY cm, effective dose 1.8074mSv). The researchers compared the images from the two scans. The outcome was that the two images were declared good (legible). The main aim of the researchers is to find if lower CT doses can be used in COVID-19 diagnosis considering its adverse effects.
Further analysis of the paper for internal and external validity, alongside statistical significance shows that the paper is at a high risk of bias as the modalities for recruitment of the participants was not explained. Also, reporting that the CT-Scans were `good` requires further description;
`good` is vague. The authors of the study did not say if there was any effort to control confounding factors. The sample size is too small to make for significance statistically. The study has to be repeated elsewhere under the same conditions before generalisation and application can be done.
Trials on Molecular Tests and SARS-CoV-2 Receptors
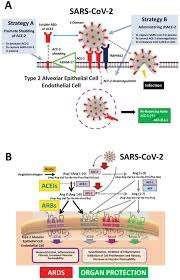
Figure D- SARS-CoV-2 Binding to ACE 2 Receptor
Adapted from Rossi et al., 2020 [Available at http://www.google.com; accessed May 15, 2020 1343 hrs GMT]
Four of the nonrandomised trials reviewed are on the receptors on which SARS-CoV-2 bound to infect the cell. One study was on modifying the traditional receptor, the ACE II receptor by crystallization (Shang et al., 2020). Another was on integrins which some authorities have posited as an alternative receptor for the SARS-CoV-2 to gain access to the human cell (Sigrist et al., 2020). One other studied E gene as an enhancer for the detection of RdRp antigen protein in the diagnosis of COVID-19 pneumonia (Konrad et al., 2020).
In addition, the fourth study looked at the interaction of SARS-CoV-2 and ACE II receptors, angiotensin II blockers and RAS inhibitors (Kai H, Kai M, 2020). The study was done in Japan. Preliminary results of research done in mice revealed more severe lung injury in mice which have had their ACE II receptor knocked out.
Similarly, another study mentioned that hypertensive patients on ACE inhibitors and/or Angiotensin Receptor blockers had a 6% absolute risk reduction in risk of contracting COVID19 when compared with hypertensive patient not currently taking the drugs, (that is, patients on
placebo). The study found out that while 9% of participants in the study who were on ACE inhibitors/Angiotensin Receptor Blockers (ARBs) were eventually diagnosed with COVID-19 at the end of the study, only 3% of participants on the ACEi/ARBs were COVID-19 positive at the end of the study (Number Needed to Treat=33).
The study on crystallisation of the ACE II receptor reported that the receptors were easier to study after crystallisation. The exact process of the crystallisation process was not clear. The recruitment criteria were not given as well. Going further the internal and external validity as well as statistical significance cannot be assessed. Similarly, the second study reported that RGD motif (part of the spike protein of the virus) binds to an integrin that is within close proximity to ACE II receptor. By so doing, the researchers posited that the integrin can be an alternative route of entry of the virus (Sigrist et al., 2020). This warrants more research. The actual values of the study were not given, therefore, assessment for bias, confounding risks, error and generalisability could not be done.
Another paper on receptors was done in mice and actually studied the SARS-CoV, the pathogen of Middle East Respiratory Syndrome (MERS). The researchers compared mice that were given an ARB compared with ones that did not get an ARB. They reported that the mice that got an ARB suffered less lung injury compared with ones that did not get. The researchers recommended continued use of ACEi and ARB in hypertensive COVID-19 patient until future research disproves their use. Further information was not given. The values (results) of the study were not made available.
The study was on mice and cannot be applicable to humans until done in humans. Secondly, there is a high risk of selection bias and hasty generalization. The study was done on the MERS virus and not the SARS-CoV-2, COVID-19` s pathogen. The two viruses may be similar but are not the same. Further research is needed in this area.
Three studies on genomic analysis of SARS-Cov-2, silico assessment of the genome of SARSCoV-2 and transmission of COVID-19 in an asymptomatic contact in Germany (Stefanelli et al., 2020), (Holland et al., 2020) and (Rothe et al., 2020) were reviewed. Stefanelli and his colleagues looked at two SARS-CoV-2 strains from two patients in Italy in January and February
2020. They looked at the genomes of the two strains. They found out that the two strains were phylogenetically different.
Further assessment for internal and external validity, statistical significance in addition to chance of error revealed that the risk of bias is high considering the participants were inpatients. Also, the small sample size increased the likelihood of the result being due to chance and therefore reducing the statistical significance of the study. The study has not been repeated, hence its results cannot be generalised.
Holland and his colleagues did a study similar to the one done by Stefanelli and group. However, the Holland group studied the SARS-CoV-2 in greater detail. The Holland group identified 25 disparate sequence regions on the virus. The outcome of their study showed that they could identify only four out of the 25 targeted sequences in their samples. The study is at a risk of bias as the recruitment of the participants was not described. Statistical significance was not provided and the p-value or any other value was not available. Findings from the study cannot be generalised as the study has not been done anywhere else.
The third group, Rothe and colleagues did a study almost identical to the work of Stefanelli`s group. They studied the SARS-CoV-2 strains from two COVID-19 (an Italian and a Chinese), both males and asymptomatic. The research work was done in Germany. After analysing the genomes of the strains, they found that the Chinese patient had a strain `native` to China while the stain from the Italian was almost identical to the other ones found in Germany. They then concluded that the strains that caused the infection in Germany were not entirely from China. This agrees with a research finding done in The Gambia which traced the SARS-CoV-2 strains in The Gambia to Japan; and United Kingdom and Spain (Europe) (Kanteh et al., 2020).
Further review for internal validity reveals high risk of bias as the recruitment process of the participants was not defined. Confounding factors were not controlled. Though sample size of 284 isolates look fairly sizable, the researchers did not mention how many persons the samples were got from. No p-values or confidence intervals were given, hence, statistical significance
cannot be determined. The study has to be repeated under the same conditions elsewhere before the results can be generalised.
Clinical Trial on Saliva
A paper on the value of saliva in COVID-19 diagnosis was reviewed (Xu et al., 2020). The researchers collected `coughed up` saliva (n=12), saliva swab (n=23), salivary gland swab and oral cavity (n=15); *n is number. The researchers registered varying sensitivities of 12.9% (salivary gland swab), 50% (saliva in the oral cavity), 86% (saliva swab) and the very high sensitivity 91% in `coughed up` saliva.
For internal validity the risk of bias and efforts to control for confounding risks were evaluated. The risk of selection and observer bias is high. The criteria for recruitment for the study were not given by the researchers. The researchers did not specifically mention if asymptomatic patients were recruited in the study. Otherwise, it is presumed that they are all inpatients. This will increase the risk of bias. Confounding factors could have been better controlled by using agematched controls. The controls should also share similar sex and medical comorbidities as the patients. This was not done as the researchers did not mention it.Statistical significance could not be measured as the p-values and confidence intervals were not provided by the researchers. External validity could not be guaranteed as the study was done in a hospital setting and until it is repeated elsewhere the findings are not generalisable.
Clinical Trials on Artificial Intelligence
A nonrandomised trial on the CRISPR-based deep learning programme that was done in the US in 2020 was reviewed (Metsy et al., 2020). The population studied were viral assays .The intervention done was applying CRISPR-based deep (machine) learning to detect pathogens including SARS-CoV-2. The researchers compared the result with the results from nucleic acid amplification (RT-PCR). The outcome is detection of 67 viruses including SARS-CoV-2. The purpose is to develop rapid diagnostic tools to detect viruses.
Further analysis to test for internal and external validity, as well as statistical significance. I found out that the study was at a high risk of bias as the source and method of extraction of the assays were not given. Also, no p-value or confidence interval was given. The likelihood of the
findings being as a result of chance is high. In fact, the authors of the paper gave only a qualitative analysis and no quantitative analysis of the values. The study has not been done outside the US and thus cannot be generalised or applied in other settings across the world.
Clinical Trial on Serological Tests
A study first done in 1984 looked at the antibodies formed against the 229 E-related strains of virus (Reed SE, 1984). A group of volunteers had swabs taken from their nasal and nasopharyngeal cavities. All had viral pneumonia. There was no comparator. The outcome of is identification of antibody to the 229E antigen in 89 % of the samples while 11% were negative. The purpose of the study is to identify antibodies to coronavirus antigens.Further appraisal for internal validity showed high risk of bias as patient characteristics and health status were not explained. No controls against confounding factors were done. No quantitative analysis of the result was given; therefore statistical significance cannot be determined. It cannot also be known if the study results are by chance as the p-value was not made available.
The study was done with viruses other than SARS-CoV-2 and has to be repeated with SARSCoV-2 isolates under the same conditions before the result can be generalised.
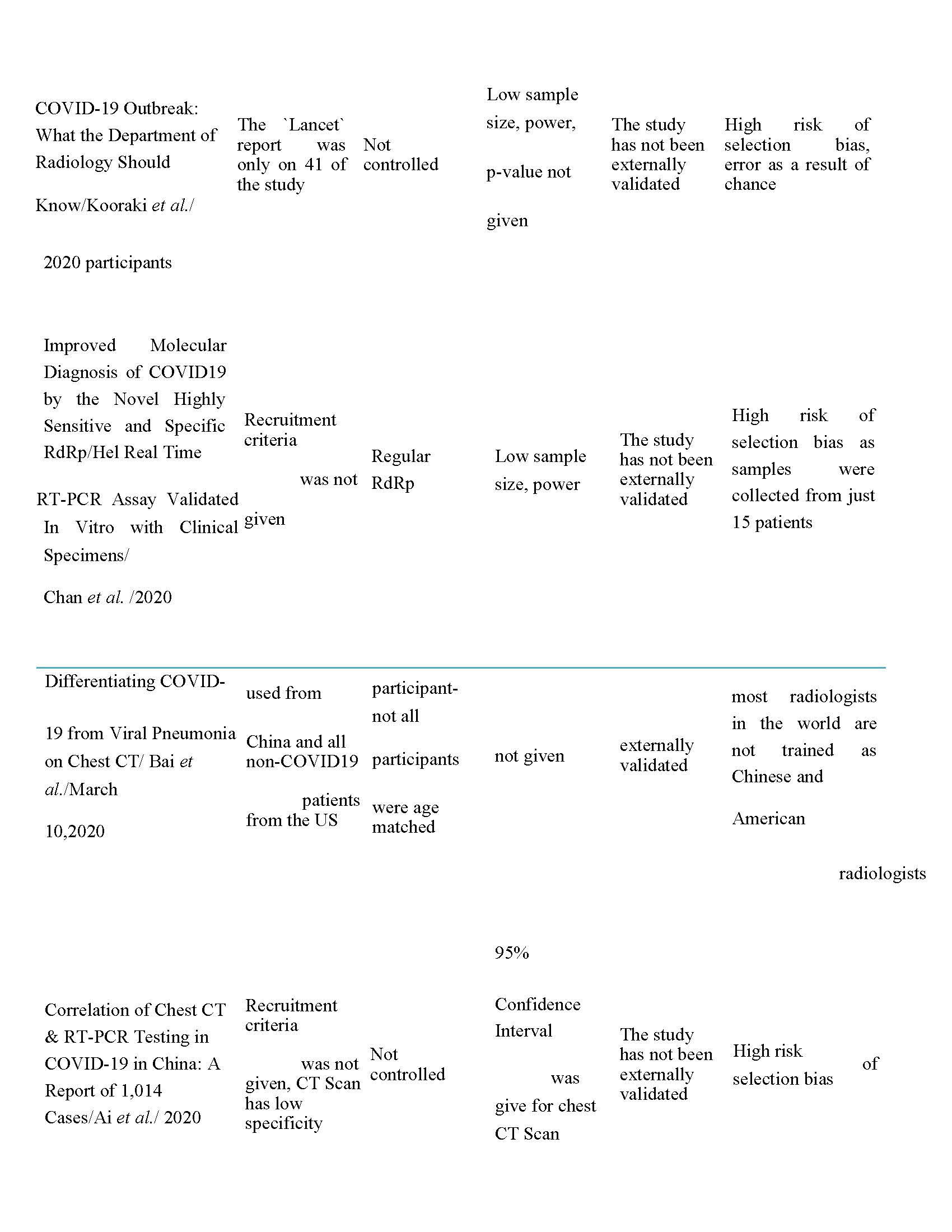

Most the reviewed papers have sample sizes that could not guarantee statistical and clinical significance. The papers were hastily done probably due to pressure on the scientific community to control the COVID-19 pandemic. The studies were biased as it appears only the severely sick patients were recruited for the studies and the ones with mild symptoms were left out. Wynants and colleagues utilised the PROBAST tool to assess for risk of bias and a good number of the models showed high risk of bias. They, however, did not show these risks were calculated.
The effect of confounding factors was not controlled with matching. Only one study included matching but not for the entire study (Bai et al., 2020). Confounding factors appear like the subject of interest-COVID-19 pneumonia and will influence result unless their effects are controlled by matching, stratification, and multivariable analysis. Many reviewed studies did not provide p-values. This is most likely linked to the low sample sizes of the studies. Some others also gave sensitivities and specificities without giving confidence intervals. These detract from statistical significance and reliability of the studies.
Almost all the studies were done in the COVID-19 hotspots of China, the US, Gernany, Italy and Japan. The findings of these studies cannot be generalised unless they are repeated in several other places across the world under identical experimental conditions. An example of the type of external validity required to lend more credence to COVID-19 diagnostic tools studies is as given by Riley RD and colleagues (2Riley et al., 2020).
Of the 20 papers reviewed, two are `rapid` reviews, two randomised clinical trials and sixteen nonrandomised studies. The first review on prediction models was on models that predicted COVID-19 likelihood of being admitted in hospital, diagnosing COVID-19 pneumonia and
detecting the prognosis of a COVID-19 patients. This review concentrated on models that first two classes of models and did not look at models that deal with prognosis in detail. The models on likelihood of hospital admission were done with small sample sizes and there is a high risk of over fitting
(1Riley RD et al., 2020); and the models on detection of COVID-19 are at a high risk of bias particularly selection bias as detected by the PROBAST. Even though the researchers gave the result of the assessment of the models with PROBAST, they did not outline how the different risks of bias were calculated (see tables 1-4 on pages 27-28).The high C Indices and perfection discrimination posted by the authors appear too optimistic. These models should only be applied clinically after further research has been done with larger sample sizes and in places other than the hotspots of China and the US where most of the studies were originally done.
The study by Chan and group on RdRp/Hel (RT-PCR) test posted promising results. However, the small sample size and the absence of controls detract from the results. Collecting 273 samples from just 15 patients makes whatever result arrived at at the end of the study not too reliable. Also, the study is at a high risk of bias as very sick patients were recruited for the study. The criteria for recruitment were not made available (inclusion and exclusion criteria were not given).
The study by Bai and his colleagues shows how useful chest CT Scans are to diagnosis of COVID-19. The researchers arrived at high sensitivities for chest CT Scan in COVID-19 diagnosis. This is in agreement with results other researchers have posted (Ai ,et al., 2020).Chest CT Scan have been known to give high sensitivity but low specificity as the other viral pneumonia also present with the classic `ground glass opacity`. Bai and his group went further to show that radiologists can give CT Scan which hitherto had low specificity some shot in the arm as regards specificity. The end result is that Chest CT Scan, alongside interpretation by experienced radiologists will now have both high sensitivity and specificity for diagnosing COVID-19.
However, the study by Bai and his group erred in that they used COVID-19 patients from China and non-COVID-19 patient from the US to do their study. This surely will have an impact on their results. A mix of the patients from both countries would have made the findings more credible. A cross-over study would have been good at least. The sample size of 424 is low.
The other studies on CT Scan go to show that it has good sensitivity but low specificity. The findings seem to suggest that chest CT Scan has more sensitivity than RT-PCR particularly early on in the disease before active viral shedding (Ai et al., 2020). Evidence is weak due to small sample size, selection bias, possible observer bias and low external validity (findings cannot be generalised). More research with higher sample sizes and under stricter standard operating procedures are urgently needed to validate these claims. The study should be repeated in other parts of the world to make the findings generalisable.
The studies on ACE II receptors, artificial intelligence and antibody test did not explicitly define processes of the studies (Metsy et al., 2020). They could hold diagnostic solutions of COVID-19 but current evidence do not show this. The sample size and recruitment criteria were not made available. Clinical and statistical significance can therefore not be measured.
The enormity of the disruption COVID-19 has caused all over the world has made the scientific community especially the ones working on COVID-19 the toast of the world . Any research on COVID-19 pneumonia will certainly attract funding. The researchers must make the best use of this goodwill. The results of some researches done so far are promising. They just have to be done under Standard Operating Procedures (SOPs) with larger samples sizes and fair, randomised recruitment of participants in studies-the studies should have patients of diverse health status. There should a good mix of severely sick, mildly sick and asymptomatic COVID19 patients.
Confounding factors should be controlled with age-matching, stratification and multivariable analysis. The studies should also be blinded, where possible, double blinded. There has been little or no generalisability of the findings.External validity is therefore lacking. Researchers in the future can guard against this by building global partnerships so their study findings can be peer-reviewed and repeated by colleagues in the other parts of the world.
There is also a role for serological test, that is, antibodies. They are still being developed and can help detect individuals that have had COVID-19. This knowledge can be applied in development of vaccine. Antibody tests-IgM and longer-lasting IgG are believed to confer some form of immunity on recovered patients. The pandemic is still occurring and it will take research over several years to confirm this. An application of the conferred immunity will lead to development of vaccines. It has been posited by experts that immune passport (acquiring lifelong immunity after infection) can be applied in assigning formerly infected health workers to care for currently infected patients.
Artificial intelligence is also gaining a lot of ground with regard to diagnosing COVID-19. Deep (machine learning) statistical tools are being deployed in the diagnosis of COVID19. Robots could be deployed in the direct care and contact with infected patients. All these are within realms of possibility, further research will unravel them.
The author is immensely grateful to all the researchers whose works were used for this review for making their works accessible. Conflict of Interest: The author has none to declare.
Espinoza et al. A Guide to Chatbots for COVID-19 Screening at Paediatric Health Care Facility. JMIR Public Health and Surveillance; Volume 6, Number 2, 2020 (April-June 2020)
Konrad et al. Rapid Establishment of Laboratory Diagnostics for the Novel Coronavirus SARS-CoV-2 in Bavaria, Germany. Eurosurveillance; February, 2020
Chan JF, Yip CC, Yuen KY. Improved Molecular Diagnosis of COVID-19 by the Novel Highly Sensitive and Specific 19-RdRp/Hel Real Time Reverse Transcription-PCR Assay Validated In Vitro with Clinical Specimens; 2020 [PubMed]
Kang et al. Recommendation of low-dose CT in the Detection and Management of COVID-19. European Radiology; 2020
Xia Guo Shao et al. Clinical CT Features in Paediatric Patients with COVID-19 Infection: Different Points from Adults. Paediatric Pulmonology; Volume 55, Issue 5; 2020
Ai et al. Correlation of Chest CT Scan and RT-PCR Testing in Coronavirus Disease (COVID-19) in China: A Report of 1,014 Cases. Radiology; 2020
Bai HX, et al. Performance of Radiologists in Differentiating COVID-19From Viral Pneumonia on Chest CT. Journal Radiology; March, 20200
Vaishya et al. Artificial Intelligence (All Applications for COVID-19 Pandemic). Diabetes and Metabolic Syndromes: Clinical Research and Reviews; Volume 14, Issue 4, July- August 2020, Pages 337-339
Reed SE. The Behaviour of Recent Isolates of Human Respiratory Coronavirus In Vitro and in Volunteers: Evidence of Heterogeneity Among 229E-Related Strains. Journal of Medical Virology; Volume 13, Issue 2
Wynants L, Calster BV, Bonten MM, et al. Prediction Models for Diagnosis and Prognosis of COVID-19 Infection: Systematic Review and Critical Appraisal. BMJ 2020;369:m1328
Dong E, Du H, Gardner L. An Interactive Web-Based Dashboard to Track COVID-19 in Real Time. Lancet Infectious Diseases 2020: S1473-3099 (20) 30120-1 doi: 10.1016/S1473-3099 (20) 30120-1. pmid: 32087114
Shi Y, Yu X, Zhao H, Wang H, Zhao R, Sheng J. Host Susceptibility to Severe COVID19 and Establishment of a Host Risk Score: Findings of 487 Cases Outside Wuhan. Crit care 2020; 24: 108. Doi:10.11861S 13054-020-2833-7
Lopez-Rincon A, Tonda A, Mendoza Maldonado L, et al. Accurate Identification of SARS-CoV-2 from Viral Genome Sequences Using Deep learning. bioRxiv [Preprint] 2020. doi:10.1101/2020:03.13.990 242
Meyer B, Drsoten C, Muller MA. Serological Assays for Emerging Coronaviruses: Challenges and Pitfalls. Bonn, Germany; March 23, 2014
Kooraki S, Hosseiny M, Gholamrezanezhad A. Coronavirus (COVID- 19) Outbreak: What the department of Radiology Should Know. Journal of American College of Radiology; 2020
Rothe et al. Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. New England Journal of Medicine; 2020
Xu, Cui, et al. Saliva: Potential Diagnostic Value and Transmission of 2019-nCoV. International Journal of Oral Science (IJOS); April 17, 2020
Kanteh A, Manneh J, Jabang S, Kujabi M, Sanyang B, Oboh A,, Bojang A, Jallow H, Nwakanma D, Secka O, et al. Origin of Imported SARS-CoV-2 Strains in The Gambia Identified from Whole Genome Sequence. Medical Research Council at London School of Hygiene and Tropical Medicine, The Gambia Unit. April 30, 2020
Gong, Ou, Qui, et al. A tool to Early Predict 2019-novel Coronavirus Pneumonia (COVID-19): A Multicentre Study Using Risk Nomogram in Wuhan and Guangdong, China. medRxiv [Preprint] 2020. doi:10.110112020.03.17.20037515
Huang C, Wang Y, Li X. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet. 2020; 395: 497-506
Qin, Fang, et al. 18F- FDG/ PET CT Findings of COVID-19: A Series of Four Highly Suspected Cases. 2020 [PubMed]
Shang, Gang, et al. Structural basis of Receptor Recognition by SARS-CoV-2. Nature, 2020
Sigrist JC, Bridge A, et al. A Potential Role of Integrins in Host Cell Entry by SARSCoV-
2. March 1, 2020 [PubMed]
Kai H, Kai M et al. Interactions of Coronavirus with ACE 2, Angiotensin 2 and RAS Inhibitors-Lessons from Available Evidence and Insights into COVID-19, 2020 [PubMed]
http:// www.medicalnewstoday.com, accessed May 13,2020, 0107 hrs GMT
Feng C, Huang Z, et al. A Novel Triage Tool of Artificial Intelligence-Assisted Diagnosis and System for Suspected COVID-19 Pneumonia in Fever Clinic. medRxiv [Preprint] 2020. doi: 10.1101/ 2020.03.19.2003.9099
Song C-Y, Xu J, He J-Q, et al. COVID-19 Early Warning Score: A Multiparameter Screening Tool to Identify Highly Suspected Patients. medRxiv [Preprint] 2020. doi: 10/1101/2020.03.05.2003.1906
Meng Z, Wang M, Song H, et al. Development and Utilisation of an Intelligent Application for Aiding COVID-19 Diagnosis. medRxiv [Preprint] 2020. doi: 10/1101/2020/03.18.20035816
Yu, Shao, Guo , et al. Data-Driven Discovery of Clinical Routes for Severity Detection in
COVID-19 Paediatric Cases. medRxiv [Preprint] 2020. doi: 10.1101/2020.03.09.20032219
Chen JHK. Clinical evaluation of New High Throughput Luminex Nx TAG Respiratory Pathogen Panel Assay for Multiplex respiratory pathogen Detection. 2020 [PubMed]
Metsy HC, Frejie CA. CRISPR-Based surveillance for COVID-19 Using genomically Comprehensive Machine Learning Design [Pubmed], Google Scholar, bioRxiv.2020
1Ridley, Ensor, Snell, et al. Calculating the Sample Size Required for Developing a Clinical Prediction Model. BMJ 2020; 368:m441: doi: 10.1136/bmj.m441.pmid: 3218860 [Available at http://www.bmj.com]
2Ridley, Ensor, Snell, et al. External Validation of Clinical Prediction Models Using Bid databases from e-health Record or IPD Meta-Analysis: Opportunities and Challenges [Correction: BMJ 2020; 365: 14379] BMJ 2016; 353:i3140 doi:10.1136/bmj.i3140.pmid: 27334381; [Available at http://www.bmj.com]